In 2015, Herefords Australia, in conjunction with the Animal Genetics and Breeding Unit (ABGU) and Southern Beef Technology Services (SBTS), began development of a Genomics Policy. The aim of this Genomics Policy is to develop and enhance genomic selection for Australian Hereford cattle. During this process there has been much discussion around the challenges associated with implementing genomics, and possible solutions to overcome these challenges. Given the development of a Herefords Australia Genomics Policy, it is timely to reflect on what genomics is, how genomics works and what genomics will offer the Australian Hereford breeder.
What Is Genomics?
Genomics, or genomic selection, refers to the inclusion of DNA information into the genetic evaluation program (BREEDPLAN), in this case for Australian Hereford cattle. The DNA information used for genomics will be in the form of thousands of genetic markers, known as SNPs. Genotypes for these thousands of SNPs are generated by running a DNA sample from the animal on a SNP chip. SNP chips come in different densities; for example the SNP chip might have 20,000 SNP markers (20K), 50,000 SNP markers (50K) or 800,000 SNP markers (800K).
When genomics becomes a reality for Australian Hereford cattle, Hereford breeders will be able to take a hair sample from their animals and obtain a genomic test. The DNA of the animal will be extracted from the hair sample, and the DNA run on a SNP chip. The resulting SNP genotypes will then be used, along with pedigree information and performance information, in the calculation of BREEDPLAN EBVs.
How Does Genomics Work?
Genomic selection requires a reference population. This reference population consists of thousands of Hereford animals which have both phenotypes (performance data) and genotypes. Setting up a reference population has been one of the challenges in implementing genomics in many breeds of beef cattle; there simply have not been enough animals with phenotypes and genotypes which can go into a reference population.
The second group of animals are those which have genotypes, but do not have phenotypes. Typically these are young animals which have not yet reached an age where they can be performance recorded, although any animal with a genotype but no performance data fits into this second group.
Genomic selection uses the known relationships between the phenotypes and genotypes of the animals in the reference population to calculate genomic EBVs for young animals. This is illustrated below:
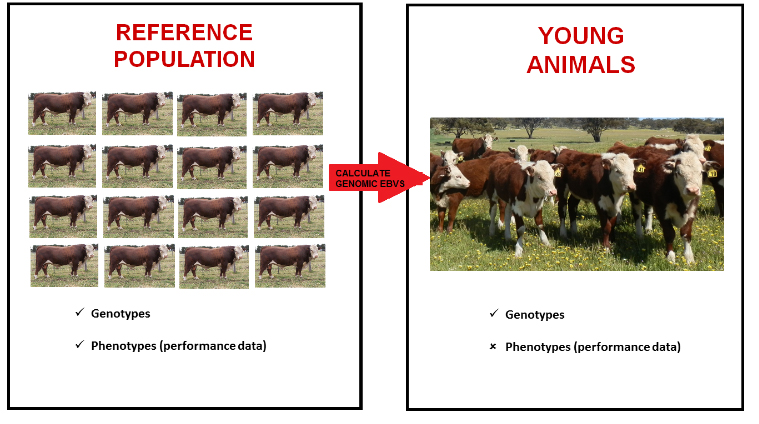
Of course, there are a number of things that influence how well genomic selection works. Firstly, the size of the reference population is important. Secondly, genomic selection works best when the reference population is closely related to the young animal population described above. For this reason, the reference population should aim to encompass as wide a range of Australian Hereford genetics as possible. This is also the reason that genomics will not replace performance recording – there will need to be new animals with both genotypes and phenotypes coming into the reference population over time. It is important that you as Hereford breeders know that genomics will not replace performance recording; the work you do to performance record your animals will be critical for the success of genomics in the future.
What Benefits Can Hereford Breeders Expect from Genomics?
Currently, the BREEDPLAN analysis uses pedigree information and performance data (both on the individual and the progeny) to generate EBVs. When genomics is implemented in Australian Herefords, a Hereford breeder will be able to take a hair sample on an individual animal, send the sample away for genotyping, and the genotype information will be included in the BREEDPLAN analysis and used to generate EBVs. This will have two main applications for Hereford breeders:
- EBVs can be generated for animals which do not have performance data There are a number of Hereford animals which are in BREEDPLAN herds, but do not have EBVs for a number of traits. This may be because:
- The animal is too young to have been measured for that trait. For example, a 200 day old calf will not have been ultrasound scanned, so is unlikely to have EMA, Rib Fat, Rump Fat or IMF EBVs.
- The trait is hard to measure. For example, Net Feed Intake – Feedlot (NFI ‐ F) is currently only measured in animals which have been through a progeny test program. Therefore, unless an animal has been measured for NFI ‐ F or has a parent which has been measured for NFI ‐ F, it is unlikely to have NFI ‐ F EBVs of its own.
In the first case, these animals currently have to wait until they are older to get EBVs for these traits. With genomics, these animals could be genotyped at a young age (e.g. at weaning), and get EBVS for EMA, Rib Fat, Rump Fat and IMF when they are still too young to be ultrasound scanned. Where a Hereford breeder wanted EBVs on stud animals for hard to measure traits, genomics would mean that relevant animals could be genotyped and EBVs generated from the genomic information.
2. More accurate EBVs can be generated for animals with limited performance information Currently, a young animal with no performance data (either of its own or its progeny) will have mid ‐ parent EBVs of quite low accuracy. With the addition of its own performance data and performance data of its progeny, the EBV may change (depending on how well the individual and its progeny perform within their contemporary groups) and the accuracy associated with that EBV will increase.
Genomics will “boost” the accuracy of BREEDPLAN EBVs; this benefit is most pronounced when the animal has EBVs with low accuracies. For example, a young animal may have an accuracy of 30% for one EBV; with the inclusion of a genomic test, the accuracy around that EBV might become 40%. However, an older animal, which might have an accuracy of 90% for the same EBV, might only have an increase to 92% accuracy for that EBV with the inclusion of a genomic test. In this way genomics can be considered similar to the addition of progeny performance data into the BREEDPLAN analysis; when the accuracy is low additional data has a large effect, and when the accuracy is high, additional data has a small effect. Of course, the amount the accuracy of an EBV increases by due to a genomic test will vary for each trait.
These genomics applications discussed above will allow Hereford breeders to identify elite bulls and heifers at younger ages than is currently possible. For example, consider an ET program where 10 full sibling bulls have been born. They are several months of age, and where EBVs are available, they have identical mid ‐ parent EBVs with low accuracy. Which one should be kept as a bull, and which ones should be castrated? This is currently a difficult decision, because the Hereford breeder has limited knowledge of the genetic potential of these young bull calves. With genomics, the Hereford breeder can take a hair sample and pay for a genomic test on each calf, and then EBVs that includes the genomic information can be generated. The Hereford breeder can now identify which of these young full ‐ sibling bull calves has the desired genetics, and castrate the rest.
As the example above shows, the power of genomics is to identify elite bulls and heifers at a young age. This allows Hereford breeders to make selection decisions at younger ages than are currently possible, and thus shorten the generation interval. In turn, shortening the generation interval should increase the rate of genetic improvement in Australian Herefords.
Summary
Herefords Australia, AGBU and SBTS have been working on a Herefords Australia Genomics Policy to investigate how to best implement genomics into Australian Herefords. There will be several benefits of genomics to Australian Hereford breeders, including the possibility of calculating EBVs on animals that cannot or have not been measured for particular traits, and also increasing the accuracy of EBVs for young animals with limited performance data. Genomics will be an important tool for Hereford breeders to continue to make genetic improvement into the future.





